Differential Gene Analysis and Pathway Analysis of TCGA Patient RNA-Seq Dataset
Design of Pipeline to Search for Possible Cancer Drug Targets via Patient RNA-Seq Analysis
Data Science Intern Project at Cowell Biodigm Co. (2020)
Project Motivation & Background:
A drug target is a specific molecule or a biological entity (ex. PD-L1) that a drug will target as a therapeutic intervention. In the drug discovery space, there are two different approaches in discovering drug targets: top-down and bottom-up. A top-down approach is where researchers start with a certain disease (ex. liver cancer) and work backwards to identify potential drug targets. If the pathology of the disease such as underlying molecular mechanisms and associated pathways is unknown, experiments must be followed to identify the key molecules and pathways that are associated with the disease. A bottom-up approach is the opposite, as it starts with a focus on the molecular components and pathways that are known to be involved in various different cellular processes. This is different, as these molecular components and pathways will be associated with many different diseases, and once a specific molecular component and pathways are defined, researchers look for possible targets for certain diseases.
Computational drug target identification has been widely used over the industry, and there are many different methods to do this. One method that is widely used (which is the method that I also used), was the phenotype-based method. Phenotype-based computational drug target identification compares the biological phenotype, which are the -omics data. Out of the four major -omics data (genomics, transcriptomics, proteomics, metabolomics), I focused on transcriptomics, or RNA-Seq data. RNA-Seq data contains many key components, but the most important component is the gene annotation and its read count. Gene annotations are essentially the labels for the gene, and allows one to identify the gene and its associated pathways. Read counts are the gene expression levels, and the higher the read count, the higher the gene expression level is. Typically, when doing a RNA-Seq analysis, data from two opposite groups are given (ex. healthy vs disease, treated vs untreated), and in my case, I looked cancer vs healthy RNA-Seq data. When given these two sets of data, the standard protocol is to first perform a differential gene expression (DGE) analysis and then a pathway analysis with the list of genes that are differentially expressed. This makes sense because the list of genes that show different level of expressions between the two group probably has something to do with the disease, and then we take those list of genes and perform a pathway analysis to see which biological pathways are associated with those list of genes.
However, it is important to capture the topology of the pathways. In a pathway analysis, the goal is to find the most statistically perturbed pathways, and then get a list of those perturbed pathways and search for possible targets. However, standard methods such as the GSEA pathway analysis ignores the topology and just regards the pathways as a simple gene set all grouped together. Pathway databases such as KEGG that is utilized here, however, contain topology in the form of graphs with nodes and edges showing interactions between genes and proteins. The comparison diagram below shows the difference between a topology-aware pathway and a simple gene set for the same pathway.
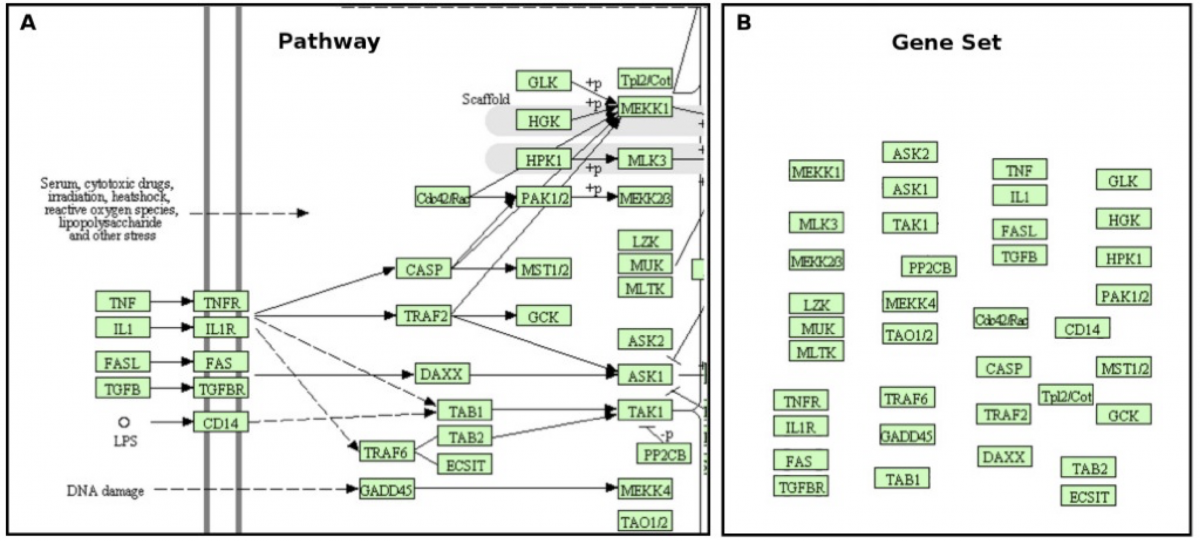
Despite correcting for multiple hypothesis testing and utilizing the topology of the pathways, pathway analysis methods still suffer from false positives and negatives. Therefore, I utilized the “primary disregulation” method summarized in this article, which essentially reduces the false positives and negatives and boosts accuracy by “distinguishing between genes that are true sources of perturbation” (more important, e.g. due to mutations, epigenetic changes, etc) and “genes that merely respond to perturbation signals coming upstream” (less important). A numerical value of “primary disregulation” or pDis is calculated for each pathway which denotes the “change in a gene expression inherent to the gene itself”. The p-value of this pDis value, or ppDis is also calculated and the list of the perturbed pathways with a low pDis and ppDis value can be further investigated to find for possible drug targets. Lastly, a “cut-off free analysis” is hypothesized to be superior, which doesn’t perform the DGE analysis before performing the pathway analysis, as the DGE analysis usually eliminates almost 99% of all the genes available in the original RNA-Seq data, possibly removing important genes that may be relevant to the disease. Both cut-off free and cut-off analysis was tested.
Context: For some context, my research internship was at a small start-up company focused on early drug discovery. The company aimed to discover new drug compounds or new drug targets for a known compound. Then, the company would aim to out-license these early “hits” or “leads” to a bigger pharmaceutical company for profit.
Methods:
- Differential Gene Expression (DGE) Analysis:
- Use R Packages called voom, limma, edgeR for DGE analysis.
- First, normalize the read counts using voom (log of counts per million (CPM)).
- Then, perform statistical testing (eBayes, empirical bayes statistics) using limma and edgeR for differential gene expression testing.
- For cut-off analysis, cut-off DGE genes on a certain threshold. For cut-off free analysis, don’t cut-off.
- Primary Disregulation (pDis) Pathway Analysis:
- Use R Packages ROntoTools and GeneBook for pathway analysis.
- Fetch the DGE gene list and use them to perform pDis analysis (with a set bootstrap iteration), also using KEGG pathways.
- Sort the results of most perturbed pathways by pDis and ppDis values (lower the better).
Note that full code is on github.
Results:
I performed 50k bootstrap iterations of pDis analysis for both liver and pancreatic cancer (LIHC/PAAD) and then returned the list of the most perturbed pathways with low total pDis values and low p value (ppDis) values (p < 0.05). Before the analysis, a DGE analysis between cancer vs healthy patients was performed if performing cut-off analysis. However, cut-off free analysis was better, and some examples of LIHC pathways that were most perturbed were “Autophagy”, “SNARE interactions in vescular transport”, “RNA degradation”, etc. Some examples of unique PAAD pathways that were most perturbed were “Autophagy”, “Homologous recombination”, “Asthma”, etc. These names of pathways then can be searched on the KEGG pathway database to look at the associated genes and the topology regarding the pathway. Lastly, a bottom-up approach using these specific pathways can potentially lead to a new drug target.
Personal Comments:
Q: Why did I choose this project?
When I fortunately got an offer to work as a summer intern for working on a RNA-Seq pipeline, I was excited because it was my first time at not only doing an internship, but also working with RNA-Seq data and R. I was a student who was always excited about cancer research, and to be part of research that screens for possible novel targets for liver and pancreatic cancer was a fascinating opportunity I could not turn down.
Q: What did I do outside of this project?
Because it was my first time looking at RNA-Seq datasets and my first time coding in R, I did a lot of background article reading on RNA-Seq dataset and related R libraries mentioned above to perform the DGE and pathway analysis. Furthermore, preprocessing the RNA-Seq dataset also required me to learn R more, especially the packages that are often used such as tidyr, dplyr, data.table, etc.
Q: What impact did this project have on me?
This was my first experience in both working as an intern and working a computational job, so it was an eye-opening experience. I think this experience eventually shaped my future research in computational work and allowed me to find another internship opportunity regarding RNA-Seq datasets. Therefore, while the project itself wasn’t anything groundbreaking in itself, the impact was definitely long-lasting in that it got me into the industry and the field.
Image credits to: