2/3D Semantic Segmentation of Skin H&E Tissue Images
Segmentation of Skin H&E Tissue Images to Analyze Novel Cellular Biomarkers of Aging
Master's Thesis Project (2023 ~ Present)
Project Motivation & Background:
Let’s play a simple guessing game: Look at the pictures below. Can you guess who is older?


Pretty obvious, right? The man on the left is older and the man on the right (Tom Brady, if you didn’t recognize him) is younger. Now, assume we were able to receive the skin tissue samples of both men, cut them into serial sections, applied H&E stain to them, scanned/digitized them, and then looked at one part of the whole slide images (WSI). Two pairs of pictures at different magnifications (4x and 20x) are shown below, and each man’s H&E image pairs are either on the left or the right. Can you answer the same question now?
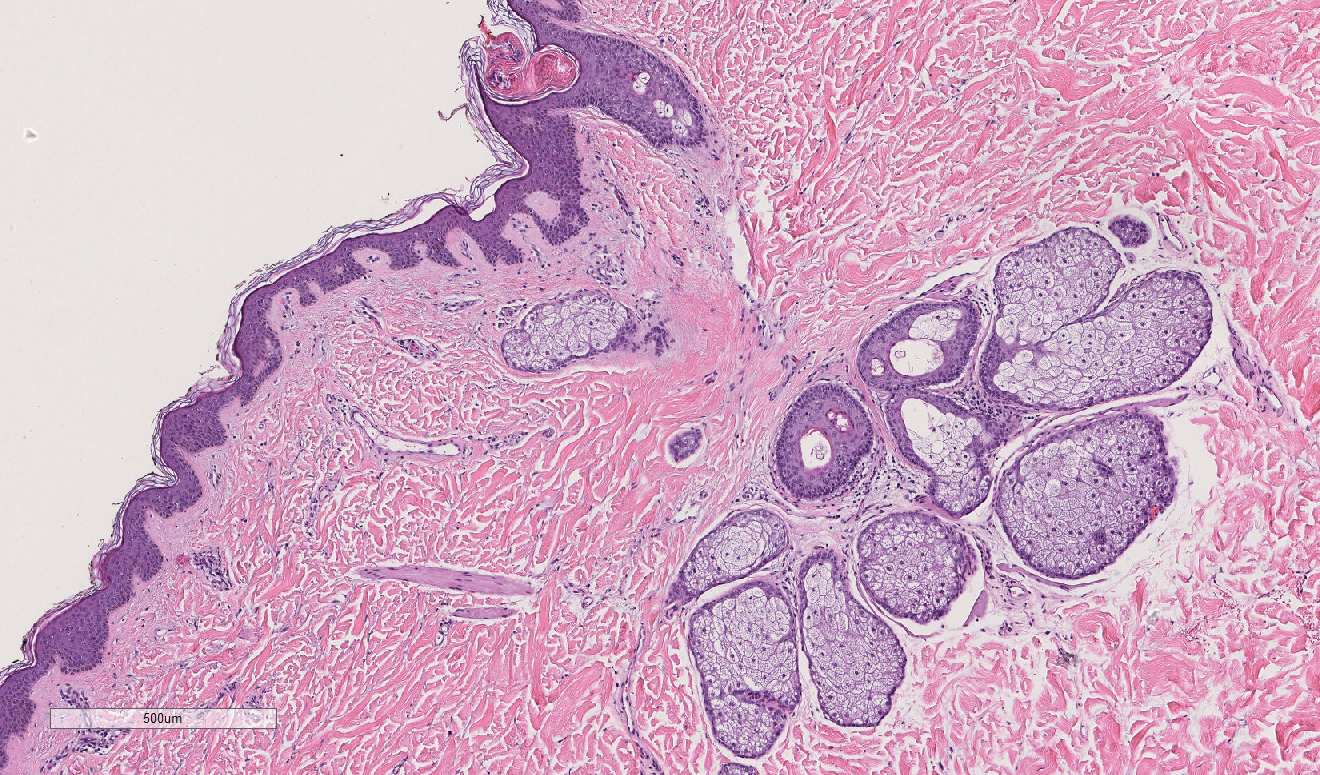
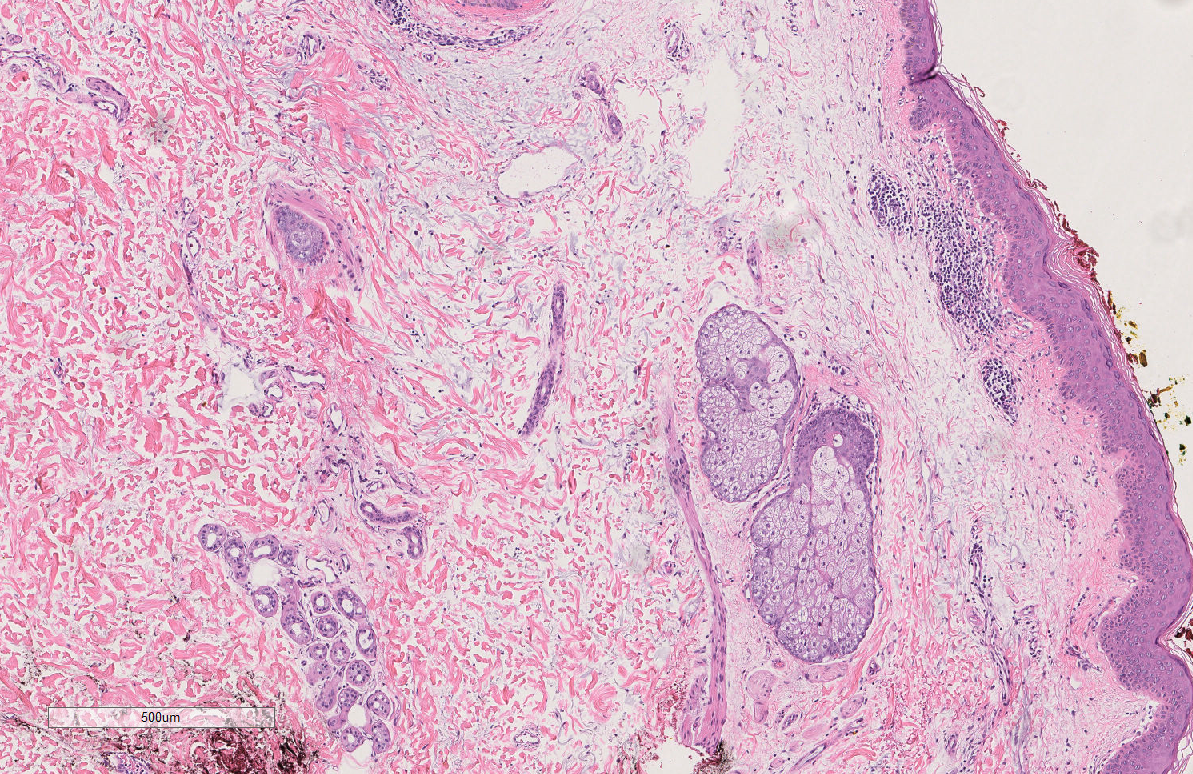
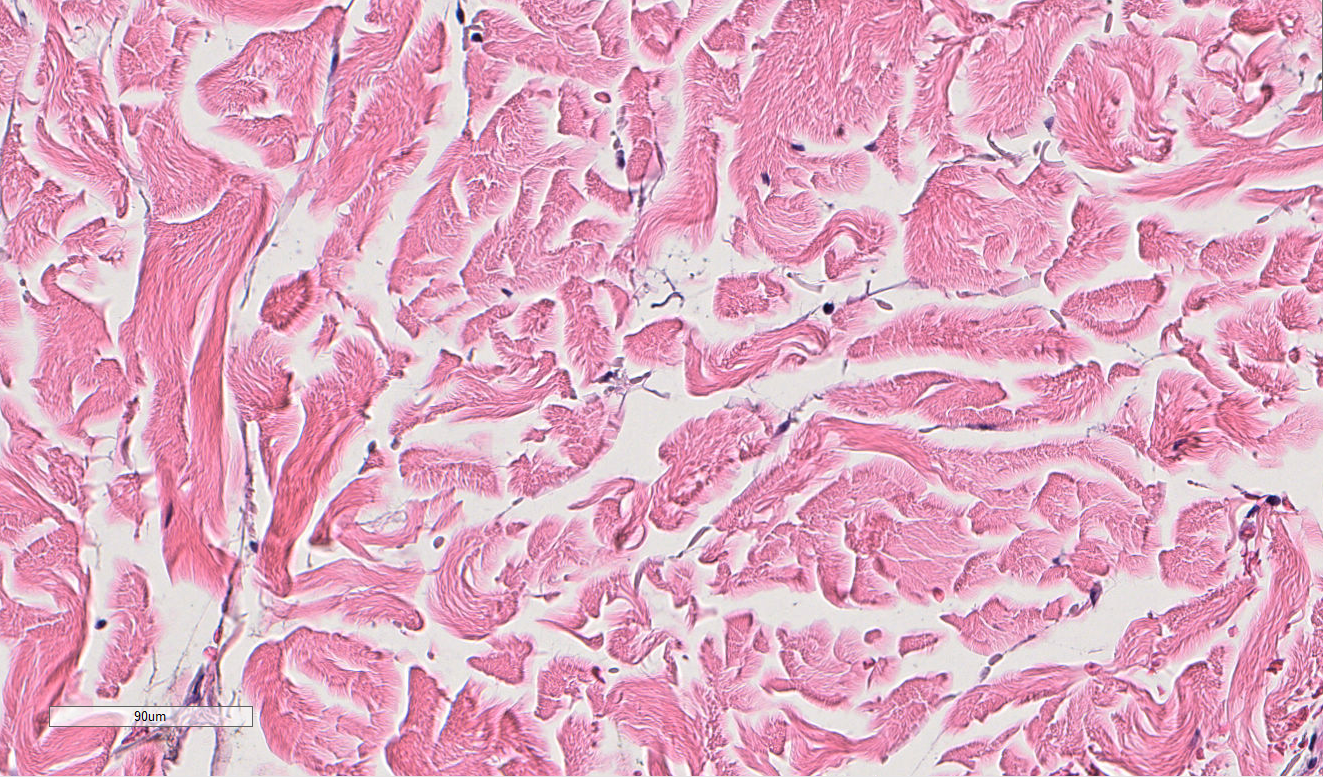
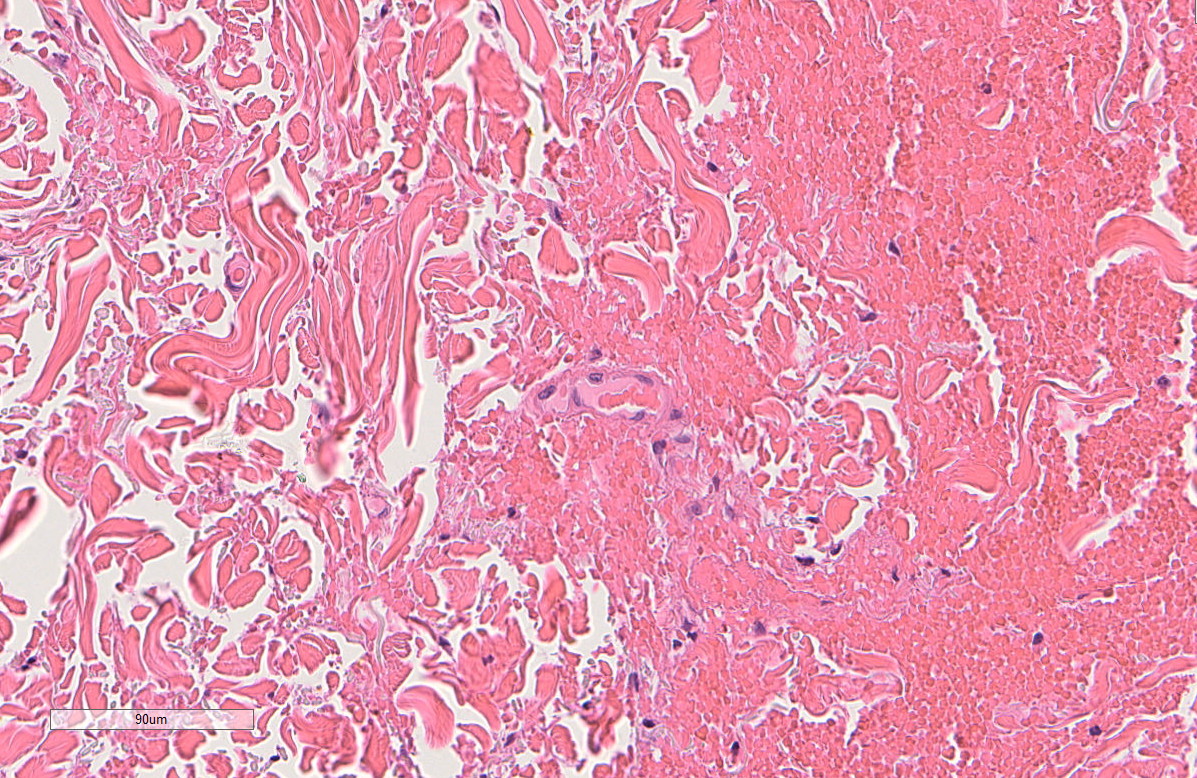
The answer is: 44-year old Tom Brady is on the left, and 95-year old man is on the right. While these tissues are not actually from the people pictured above, they actually are tissue samples from patients who are 44 years old and 95 years old. How can we tell? The picture in the first row actually shows that there are more oil glands in the left than the right, and the second row shows more ECM alignment. However, without this previous knowledge, the point is, compared to just looking at someone’s face, it’s much harder to determine a person’s age by just looking at their skin H&E tissue sample. But someone may ask, why do we want to do this? Why study aging, specifically in skin H&E tissue samples?
It’s a well-known fact that old age is correlated with virtually every disease that are the leading causes of death- heart disease, cancer, stroke, Alzheimer’s, and recently, COVID-19. To help facilitate possible interventions to prevent these chronic age-related diseases, it is critical to develop methods to accurately calculate one’s biological age. And as we’ve seen above, the easiest way of telling the approximate age of someone is to look at their face, or skin. Therefore, this study aims to find features related to aging in the skin, and like any other organs, skin function also deteriorates with age. For example, as we age, the skin loses its structural integrity and self-renewal capabilities, and experiences increased inflammation and poor temperature regulation. Look at the below diagram:
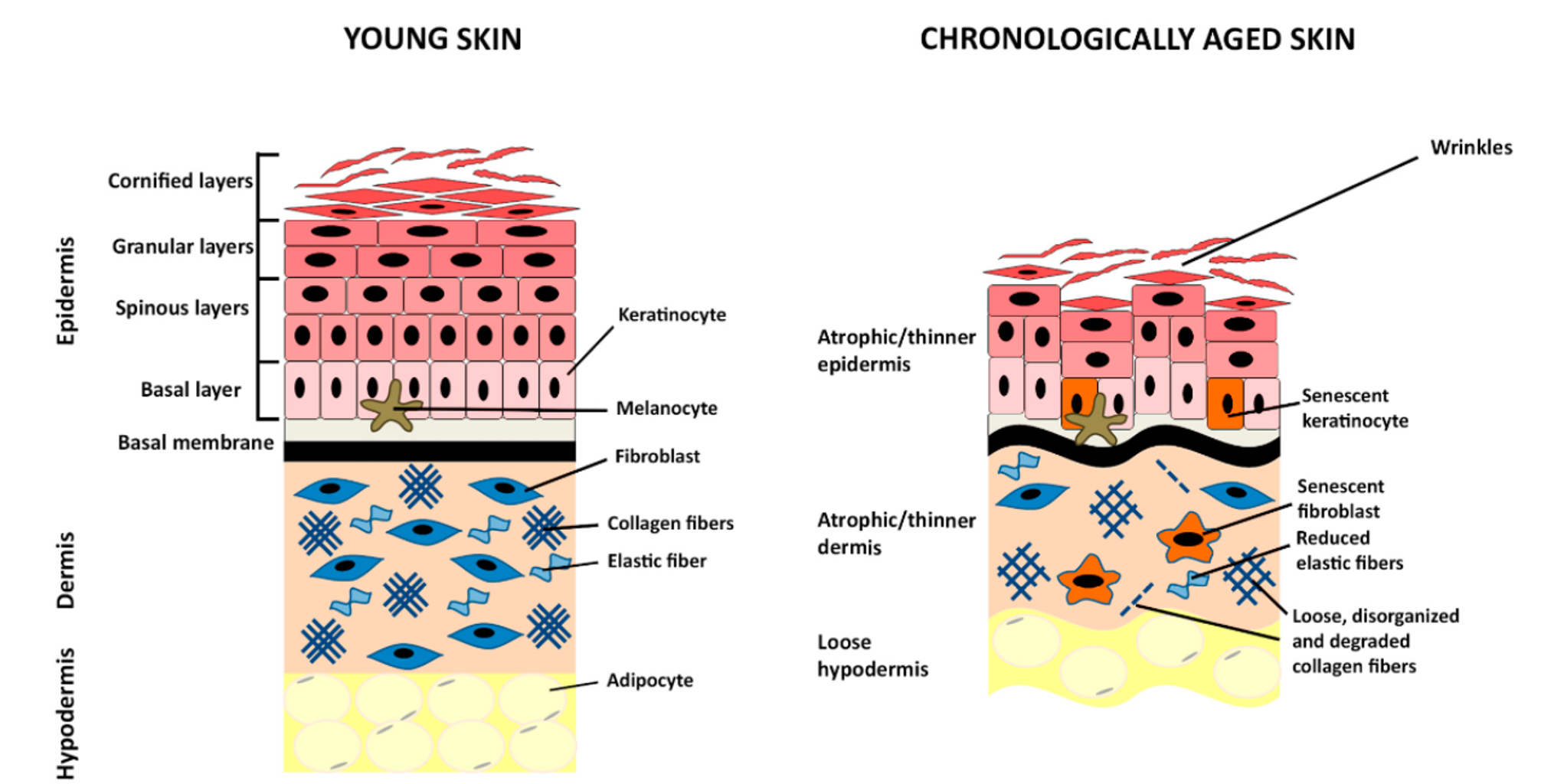
As seen in the above diagram, we already know the major differences between young and old skin. However, there has been lack of research done in the biomarkers of aging at a cellular level. Previous research in our lab has revealed biomolecular and biophysical biomarkers at the cellular level by analyzing primary dermal fibroblasts. The purpose of this study is to extend the results of this study in finding specific morphological biomarkers correlated with aging by utilizing skin H&E images to shed more light in the biological aging process.
Methods:
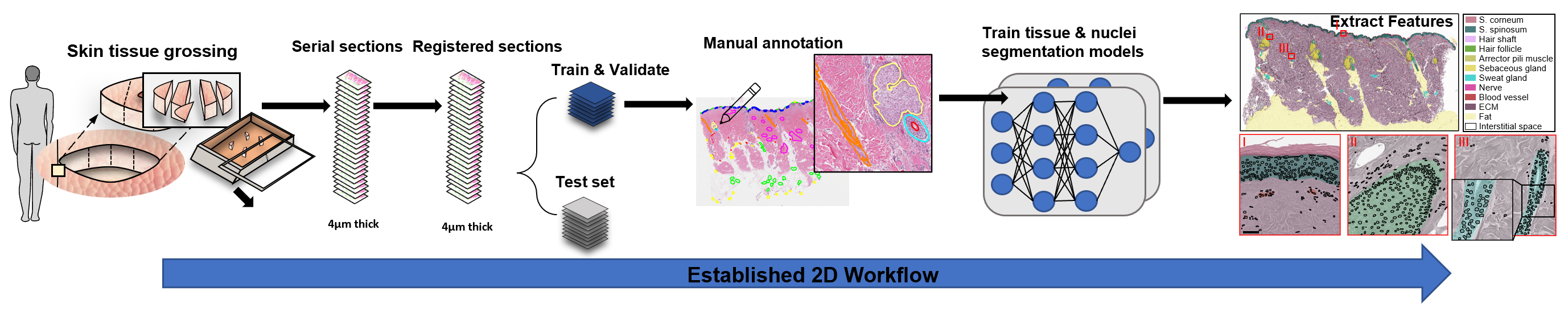
As seen in above diagram, we first obtain skin tissue samples, cut them into serial sections, apply H&E stain, and scan and digitize each slide to obtain a z-stack of 2D WSIs. Then, we undergo image registration to registrate all of the z-stack together and divide the images into a train-val-test split.
We train two different models: 1) Nuclei segmentation model using HoVer-Net and 2) Tissue segmentation model using DeepLabV3+ and/or UNet++. In order to train these supervised models, we manually annotate the nuclei or the twelve different tissue classes in the training and validation images. I hypothesized DeepLabV3+ to be a more lightweight model due to its ASPP (atrous spatial pyramid pooling) module and depthwise separable convolution, while Unet++ to be a more heavyweight but accurate model due to its dense skip connections. However, both models had negligible differences in their F-1 scores on the validation set.
Then, after confirming the model performance using overall precision and recall (F1 score) on the test dataset, we use a post-processing pipeline to extract different meaningful 2D features. Some example features to extract are individual tissue compositions and its area, distance between different cells and tissues (ex. distance between fibroblast and sweat glands), and more.
Results:
Since this is currently a work-in-progress, updates will be made whenever possible.
In total, 1090 2D features were extracted. Out of the 1090 features, through statistical testing with univariate analyses (multivariate analyses will be done in the future as well) and correlation coefficient calculations, features that were most predictive and correlated with age were selected. Additionally, gender differences and body part differences (skin from different body parts) will also be analyzed. Currently, the results are being finalized and I plan to update this soon.
Note that more technical details/explanations and further results are omitted on purpose as I focus on motivation/personal comments in introducing the project. More technical details will be shown in the technical powerpoint presentation (PPT).
Personal Comments:
Q: Why did I choose this project?
This is a fascinating project to me because it necessitates both biology and deep-learning background. I was originally planning to continue my research about gene delivery for immunoengineering, but my interests in different types of computational work grew more after my summer internship experience at Novartis. As I switched my research interests to computational work, I was initially worried because I still wanted to continue cancer research in one way or another. Also, I wanted experience in handling different types of biological/medical data. Therefore, while this project is not directly related to cancer, the project was a perfect choice because the pipeline could easily be applicable to cancer (just switch the H&E WSIs to tumor WSIs and find new ways of developing digital biomarkers for cancer instead), and handled image data, which I never had prior experience with. Therefore, since August of 2022, I switched labs and have been working on this project.
Q: What did I do outside of this project?
Since I didn't have any significant prior knowledge in the field of dermatology, histopathology, or deep learning before starting this as my Master's Thesis project, this is a fun but challenging project for me. To make up for my lack of knowledge, I always strive to do more background article reading to learn about the computational pathology space. There are a plethora of different subfields within computational pathology that interest me- for example, we can utilize various models like CNNs or ViTs for image registration, segmentation, and even translation/generation for not only H&E stained images, but also for different types of stains frequently utilized such as IHC stains. In addition, since I'm not a computer science major, I also try to continue to learn more about Python/PyTorch and probabilistic machine/deep learning everyday by taking relevant courses in school or reading online articles/textbooks.
Q: What impact did this project have on me?
This project has and will be an important turning point of my career in developing myself as an interdisciplinary scientist that is able to utilize biology/medical data and extract meaningful features out of them.
Image credits to: