Virtual Stain Conversion of IHC/Unstained to H&E Images (IHC2HE/US2HE)
Design of Generative Models to Virtually Stain/Convert IHC/Unstained (Bright-field) Images to H&E Images
Master's Thesis Project (2023 ~ Present)
Project Motivation & Background:
Let’s play a simple guessing game: Look at the two pictures below. At the most left is the ground truth IHC image to be converted. The next two images on the right are the ground truth H&E version of the ground truth IHC image and the virtually stained H&E version of the ground truth IHC image. Can you guess which is “real” and which is “fake”?
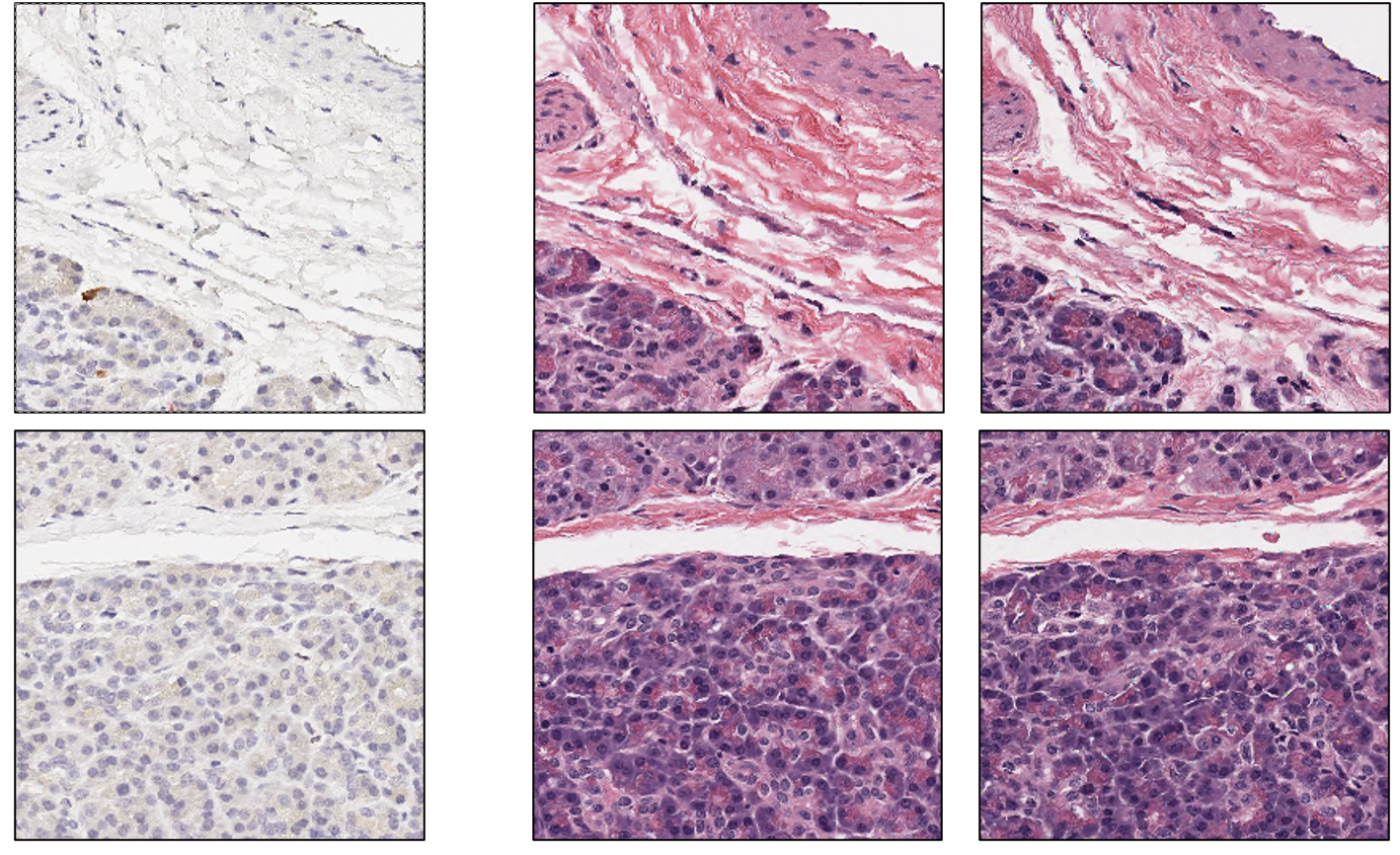
The correct answer is that the images in the right (column) are the “fake” ones, or the ones sampled from the generative model.
Let’s play another game, this time for a ground truth unstained, bright-field image to be converted. The next two images on the right are the ground truth H&E version of the ground truth unstained image and the virtually stained H&E version of the ground truth unstained image. Can you guess which is “real” and which is “fake”?
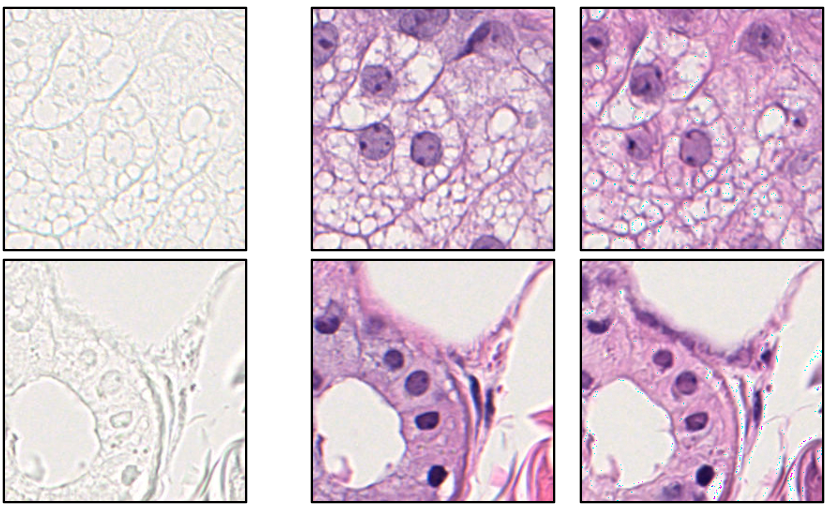
The correct answer again is that the images in the right (column) are the “fake” ones, or the ones sampled from the generative model. If you look closely, you could tell that the images on the right have slight artifacts and may be a bit “awkward” in general, this is because this is still a work in progress. With correct training, I hope to make them indistinguishable! The above two examples tell us the two different kinds of stain conversions that exist: 1) label-free-to-stain conversions and 2) stain-to-stain conversions. But someone may ask, why do we want to do this? Why virtually stain or convert stains?
First of all, commonly used histopathological stains such as H&E (Hematoxylin & Eosin), IHC (Immunohistochemistry), IF (Immunofluorescence), and others are all used to examine different types of body tissues for various research of diseases (pathology). For example, H&E and specific types of IHC stains are commonly used in cancer pathology research, as each stain visualizes the tissue and cellular structures of the tissue in its own way, and therefore allow new digital biomarkers to be developed by utilizing the stained images. However, histopathological stains are also commonly used for disease diagnostics, as for example, IHC staining for the specific tumor/cancer biomarker of HER2 can reveal different treatment plans for HER2 positive or negative patients.
While histopathological stains are widely used in disease research and diagnostics, the procedures of staining carried out in pathology labs are often time-consuming, expensive, and also can be extremely tricky to perform. The procedure is time-consuming due to the complexity of the staining process- tissue extraction, fixation, embedding, mounting, and staining are mostly all laborious tasks. Furthermore, specific types of stains, like IHC stains, can be very expensive, especially if they are used for disease research instead of disease diagnostics as obtaining large amounts of IHC-stained tissue slides can easily cost a fortune. Furthermore, the staining process itself also causes mechanical and chemical damages to the tissues, and if poorly performed, can damage the tissue. Most importantly, applying a stain on a tissue slide prevents other types of staining on the same slide, preventing further biological analysis on the same slide.
While virtual staining cannot expedite the time-consuming process of tissue extraction, fixation, embedding, and mounting, it can resolve most of the other issues mentioned above. Virtual stain conversions from label-free (unstained) slides to stained slides can not only expedite the staining process, it can be more accurate due to absence of mechanical/chemical damage on the tissue and be much cheaper as the stain does not need to be purchased. Furthermore, stain-to-stain conversions have a huge benefit as it allows staining on the same tissue slide, enabling further biological analysis on the same slide. Therefore, the main motivation of the unstained-to-HE virtual stain conversion of skin tissue images is to expedite and refine the staining process, while the main motivation of the IHC-to-HE virtual stain conversion of pancreatic tissue images is to allow multiple stain analysis on the same tissue slide.
Methods:
Below is the scheme for training and evaluating the trained model for the IHC-to-HE virtual stain conversion project. The same pipeline is applied to the unstained-to-HE virtual stain conversion project as well.
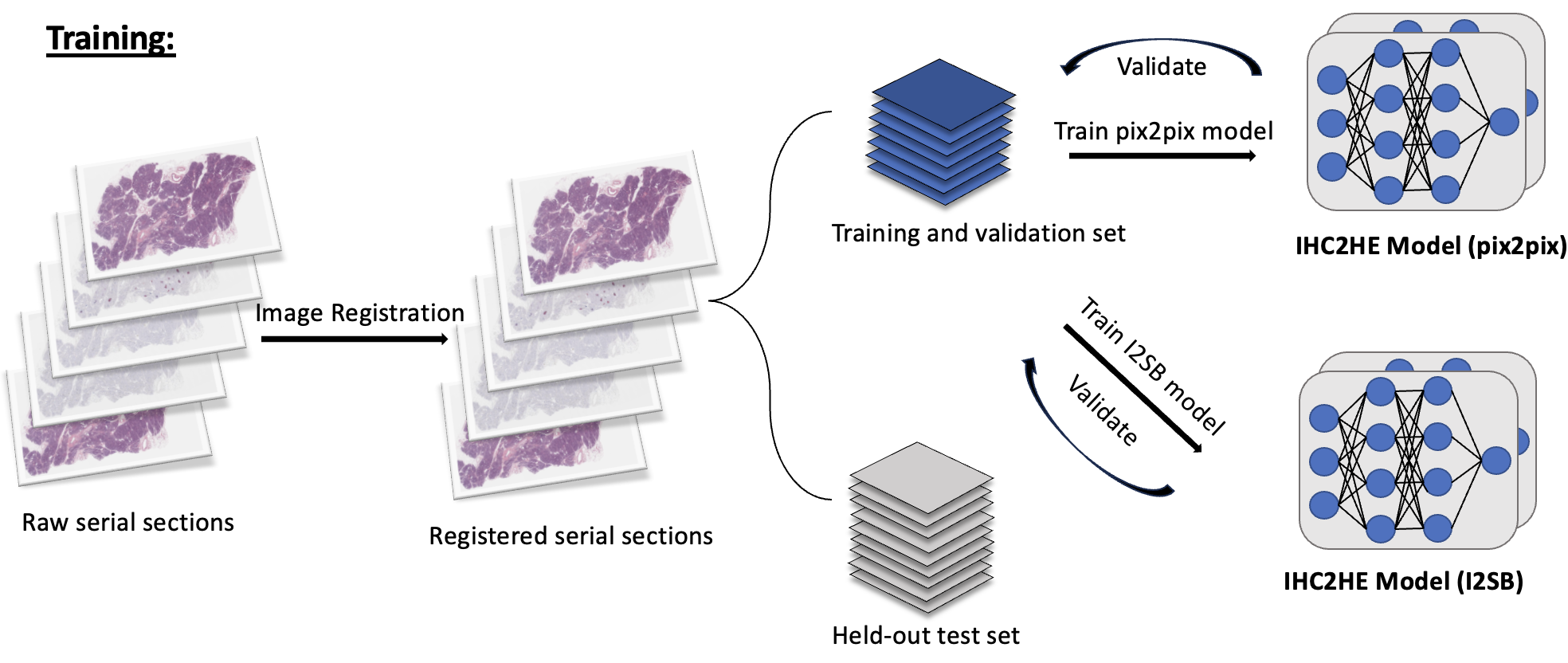
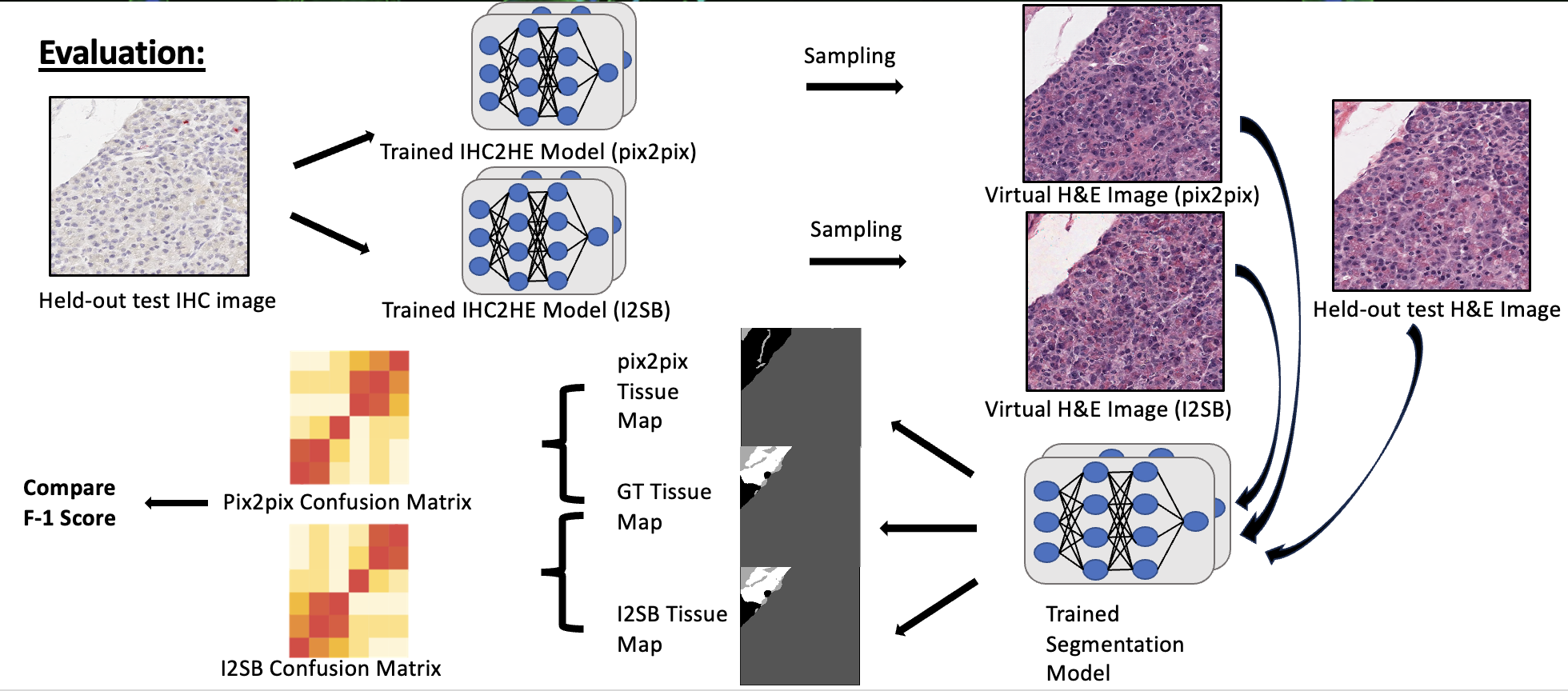
As seen above, I registered the serial sections together so that the IHC-H&E image pair are aligned properly. Then, they are tiled to 256 x 256 tiles and divided into train/val/test sets. The main thing to note here is that I am training a vanilla pix2pix (GAN) model and a more novel score-based-generative model called image-to-image schrödinger bridge (I2SB). The two generative models are trained. Then, as seen in the evaluation stage, the sampled image from the validation set is passed through a pretrained segmentation model to generate a tissue map. Next, this is compared to the ground truth tissue map generated from the ground truth image, and the F-1 score is deduced. I also plan to compare the quality of the images by calculating the inception score (IS) or/and Fréchet Inception Distance (FID) of the generated images.
To briefly talk about the models, pix2pix is a familiar model for most. Like any other GAN models, through adversarial training with GAN and L1 loss, we map the pixels of image of one domain to another. However, I2SB is more novel- it is a new class of conditional diffusion model based on training a schrödinger bridge between two different domains A and B, which finds the optimal transport path of diffusing from domain A to domain B. I2SB is related to score-based generative models, and I plan to fully explain about this in a future blog post since it is very mathematically dense. The main reason of utilizing I2SB in this project is that all of the previous works of stain-to-stain conversion out there utilize different subtypes of GANs (pix2pix, cycleGAN, etc), and do not test on the state-of-the-art diffusion models and its variants like I2SB (obviously there are more theoretical reasons to not utilizing GANs since diffusion models are known to generate more diverse images and avoids adversarial training and mode collapse). But why not use diffusion, why use I2SB? Well, look at the below diagram:
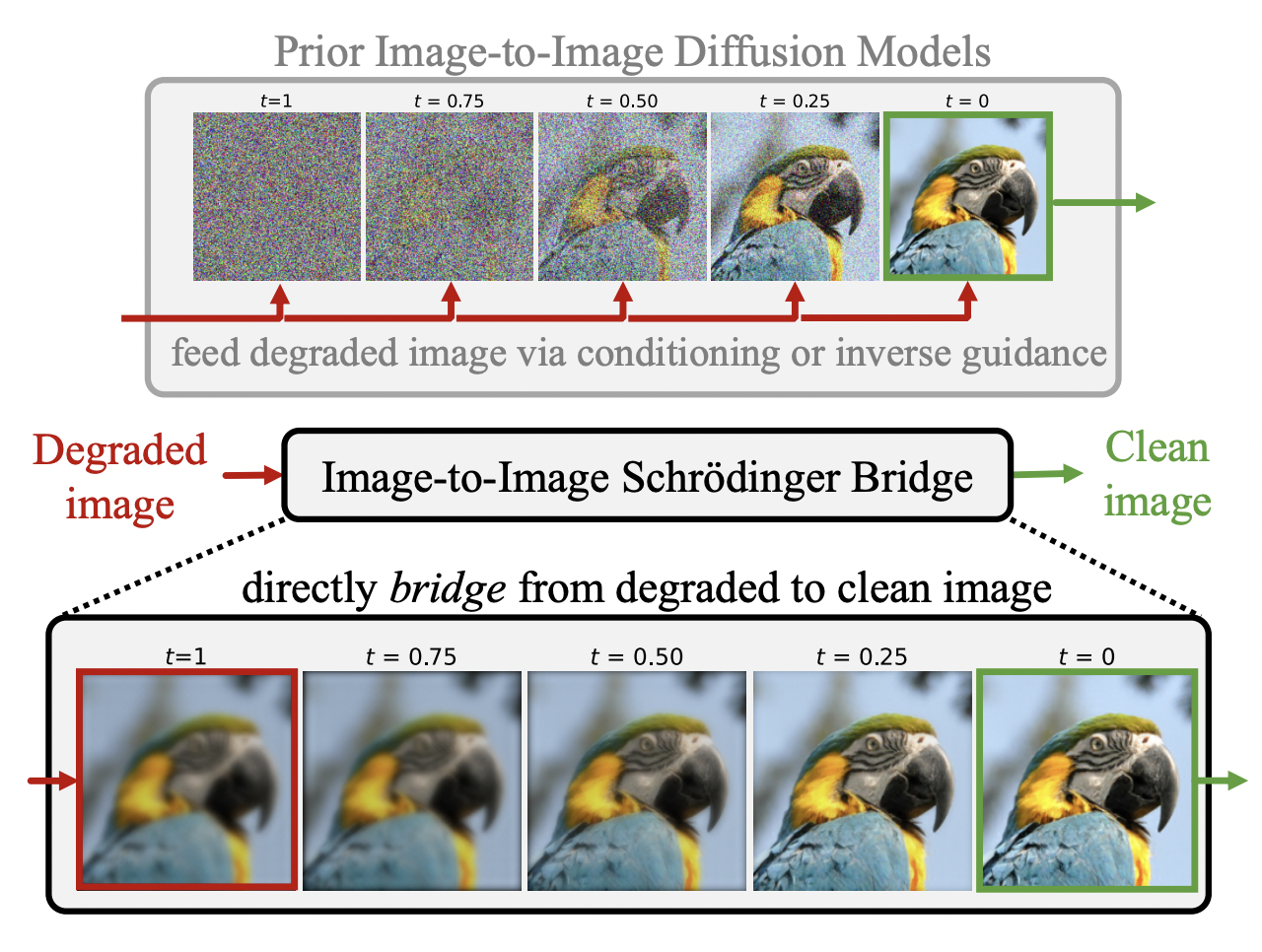
As seen above, the main difference between conditional latent/stable diffusion models and I2SB in the field of image to image translation or restoration is that the I2SB allows direct utilization of the degraded image or the image to be converted during training, which is a much more structurally informative prior compared to conditional diffusion models which start from noise during training/sampling and feeds the degraded image/image to be converted via conditioning. Likewise, in image restoration or translation, the degraded image or the image to be translated often contains useful structural information. This is definitely the case in our project, as for stain conversions, most or all of the structure should remain unchanged during the conversion. Furthermore, it also makes more intuitive sense that, to go from image A to B, it would make sense to directly train to go from A to B, not noise to B. With the models and the training/evaluation methods in mind, let’s look at the preliminary results.
Results:
Since this is currently a work-in-progress, updates will be made whenever possible.
Below are some of the pix2pix-generated H&E images compared to the ground truth H&E images:
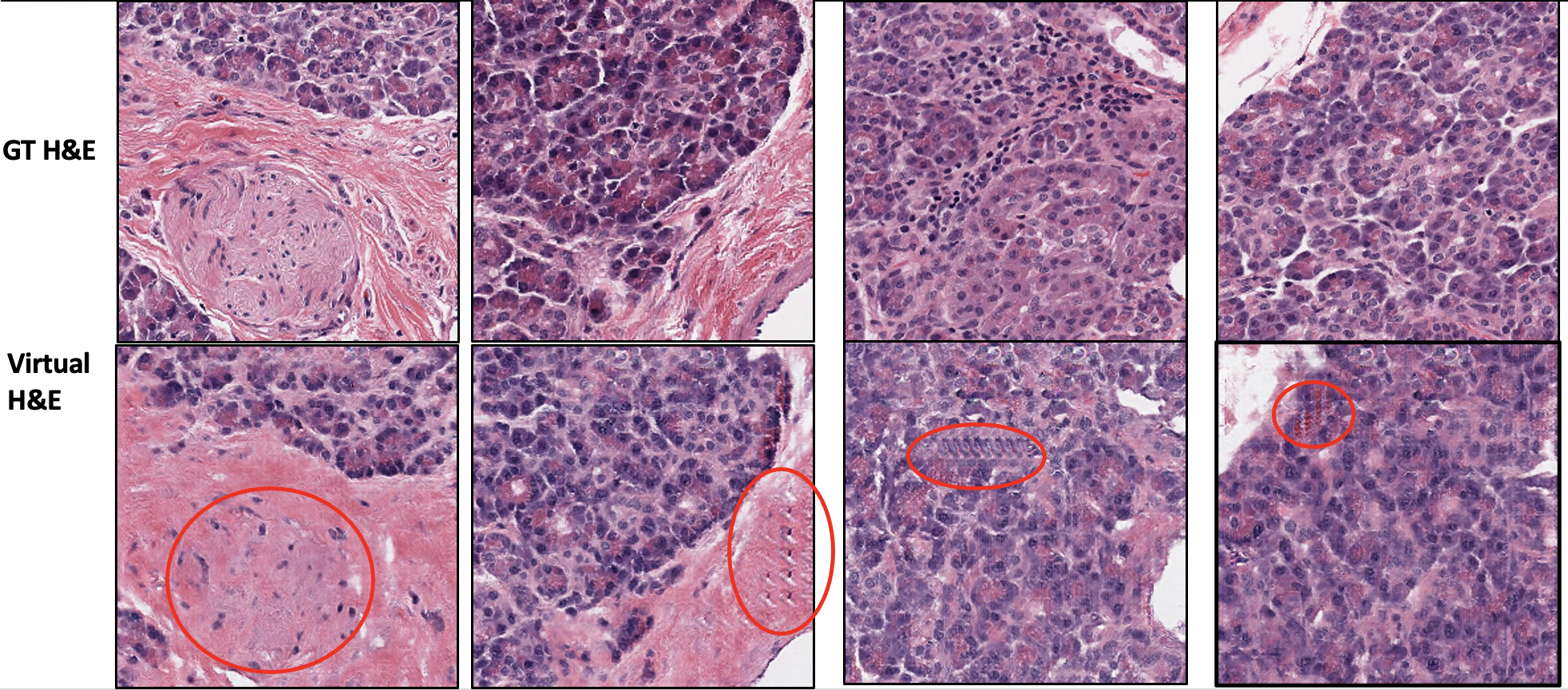
As expected for most GAN-based models, the circled region above shows low fidelity for some features of the image, image artifacts, and edge effects. Below is an example of direct comparison of pix2pix-generated and I2SB-generated image:
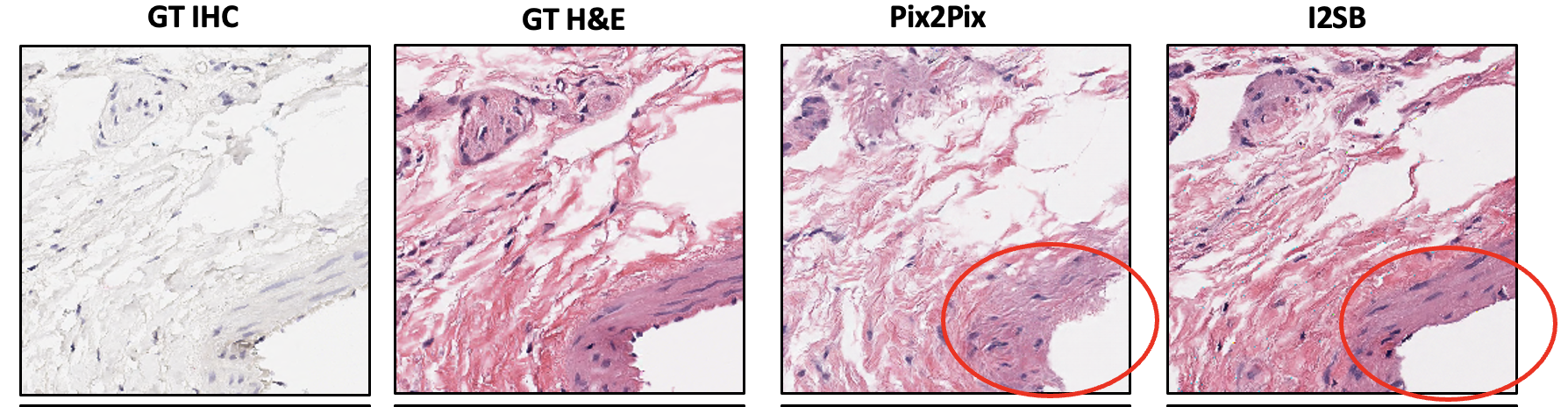
As seen in the circled part of the image and the overall image, we can see that I2SB does a better job than pix2pix in generating a more realistic image compared to ground truth. Below are some of the DDPM sampled images from the trained I2SB model:
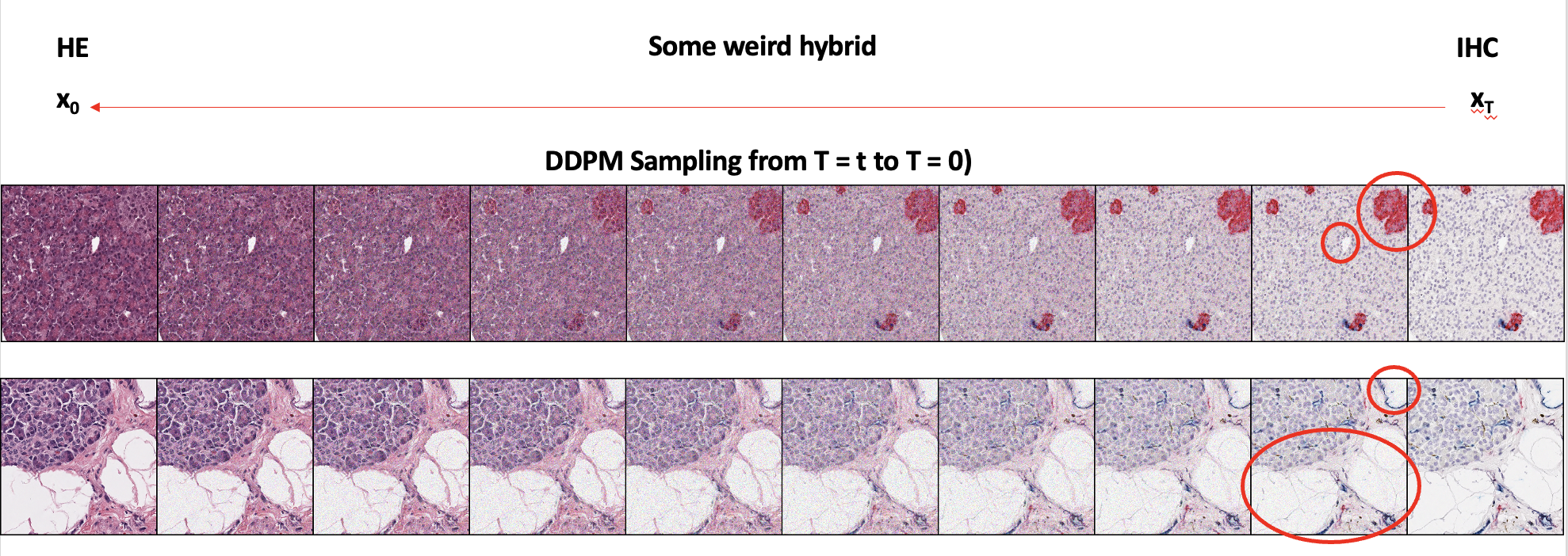
As seen in the first image, we can see that the IHC stains get properly converted to H&E as well. The second image shows local features being preserved during the stain conversion as well. With this DDPM sampling process, unlike GANs, we can also generate an interpolated image between two domains, if necessary for other uses.
The F-1 score and FID to evaluate the performance of the two models are a work-in-progress since DDPM sampling is quite slow. For the unstained-to-H&E virtual staining project, the model is still being trained, the image in the project motivation was just some image sampled from an intermediate checkpoint. As stated above, more updates will be posted here in the near future.
Note that more technical details/explanations and further results are omitted on purpose as I focus on motivation/personal comments in introducing the project. More technical details will be shown in the technical powerpoint presentation (PPT).
Personal Comments:
Q: Why did I choose this project?
After somewhat finishing training the segmentation model for the H&E image segmentation project, I wanted to independently work on a challenging project. As mentioned in the motivation above, “fortunately”, I found that our lab would benefit from a virtual stain conversion method so that multiple stains can be analyzed in the same tissue slide. Furthermore, I was excited to dive into the realm of generative models as it was very different from image classification/segmentation models that I was working on previously.
Q: What did I do outside of this project?
Similar to the H&E image segmentation project, I also had no significant prior knowledge in the field of generative models, especially on GANs and diffusion models. To utilize I2SB and understand it properly, I had to do (and still doing) a lot of intensive background research on papers for GANs, diffusion models, score-based generative models (SGM), and schrödinger bridges (SB). I’d probably say that I’ve done a lot of math learning/recap regarding statistics and differential equations during this process.
Q: What impact did this project have on me?
This project has fueled me to explore more types of deep learning fields. The field of computer vision and deep learning has still much more to offer than image classification,segmentation and generation.
Image credits to: